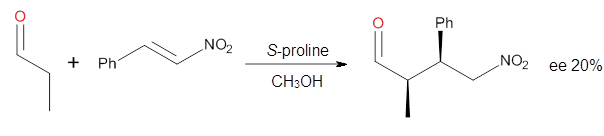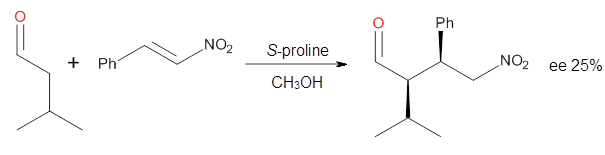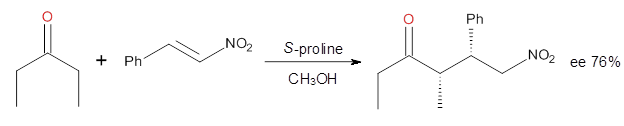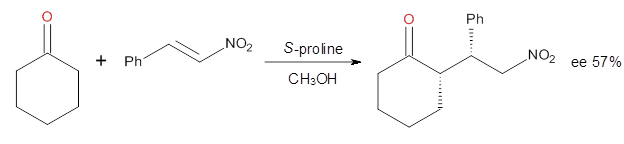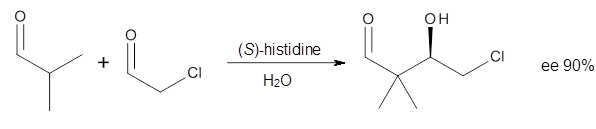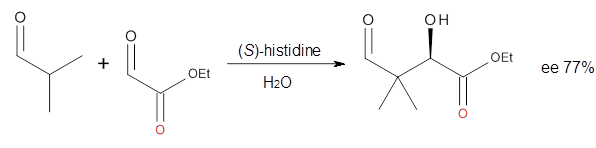Contributed by
Steven Bachrach.
Reposted from
Computational Organic Chemistry with permission
Here are a couple of articles describing computational approaches to
catalytic enantioselective reactions using variations upon the classic
proline-catalyzed aldol reaction of List and Barbas
1 that started the whole parade. I have discussed the major computational papers on that system in my book (Chapter 5.3).
Yang and Wong
2
investigated the proline-catalyzed nitro-Michael reaction, looking at
four examples, two with aldehydes and two with ketones (Reactions 1-4).
Reaction 1
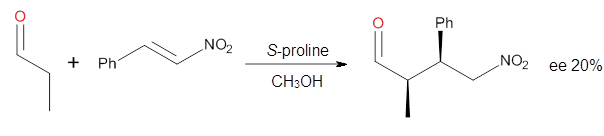
|
Reaction 2
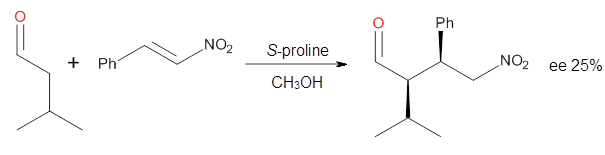
|
Reaction 3
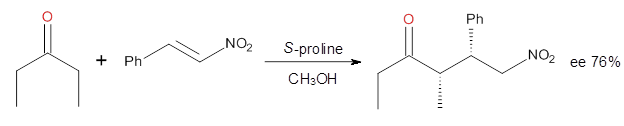
|
Reaction 4
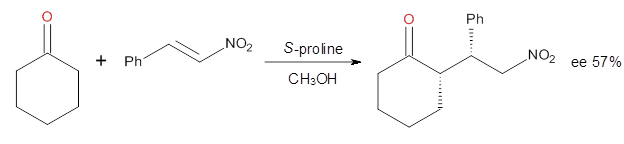
|
These four reactions were examined at MP2/311+G**//M06-2x/6-31G**, and
PCM was also applied. The key element of this study is that they
examined two different types of transition states: (a) based on the
Houk-List model involving a hydrogen bond and (b) an electrostatic based
model with no hydrogen bond. These are sketched in Scheme 1. For each
of the reactions 1-4 there are 8 located transition states differing in
the orientation of the attack on to the
syn or
anti enamine.
Scheme 1. TS models
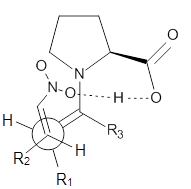
Model A
|
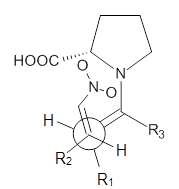
Model B
|
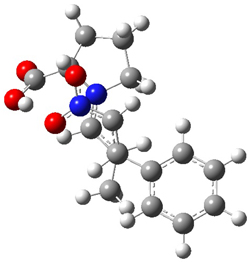
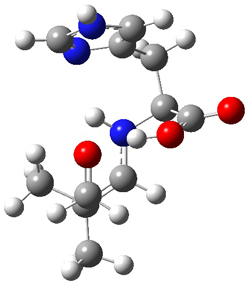
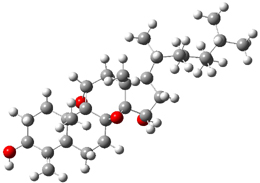
The two lowest energy TS are shown in Figure 1.
TS1-β1-RS is the lowest TS and it leads to the major enantiomer. The second lowest TS,
TS1-β3-SR, lies 2.9 kJ mol
-1
above the other TS, and it leads to the minor enantiomer. This lowest
TS is of the Houk-List type (Model A) while the other TS is of the Model
B type. The enthalpies of activation suggest an ee of 54%, in
reasonable agreement with experiment.
Figure 1. M06-2x/6-31G** optimized geometries of TS1-β1-RS and TS1-β1-RS.
The computations of the other three reactions are equally good in
terms of agreement with experiment, and importantly the computations
indicate the reversal of stereoselection between the aldehydes and the
ketones. These computations clearly implicate both the Houk-List and the
non-hydrogen bonding TSs in the catalyzed Michael additions.
Houk in collaboration with Scheffler and Mahrwald investigate the use
of histidine as a catalyst for the asymmetricaldol reaction.
3 Examples of the histidine-catalyzed aldol are shown in Reactions 5-7.
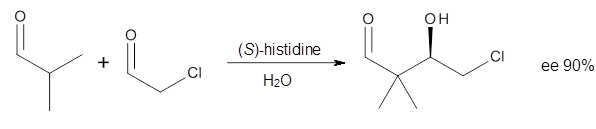
|
Reaction 5
|
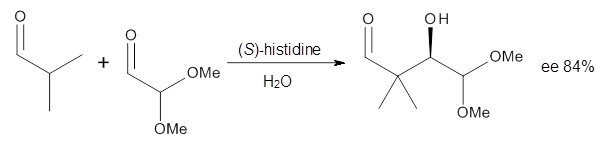
|
Reaction 6
|
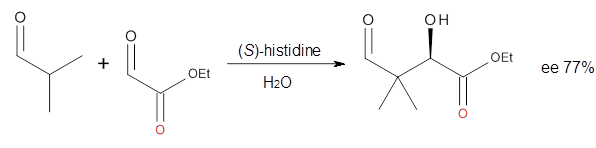
|
Reaction 7
|
The interesting twist here is whether the imidazole can also be
involved in hydrogen bonding to the acceptor carbonyl group, serving the
purpose of the carboxylic acid group in the Houk-List TS when proline
is the catalyst (Model A). The transition states for these and a few
other reactions were computed at M06-2x/6-31+G(d,p) including with the
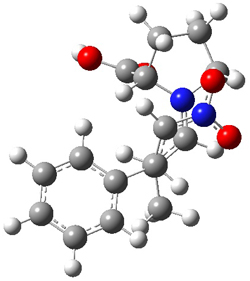
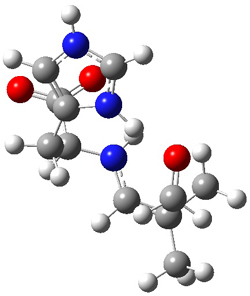
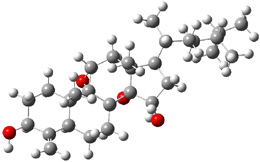
SMD continuum solvent model for water. The two lowest energy TSs for the
reaction of isobutyraldehde and formaldehyde are shown in Figure 2;
TS1 has the carboxylic acid group as the hydrogen donor while the imidazole is the donor in
TS2.
Of note is that these two TS are isoenergetic, indicating that both
modes of stabilization are at play with histidine as the catalyst.
Figure 2. M06-2x/6-31+G(d,p)
geometries of the two lowest energy TSs for the reaction of
isobutyraldehde and formaldehyde catalyzed by histidine.
The possible TSs for Reactions 5-7 were also located. For example,
with Reaction 5, the lowest energy TS involves the imidazole as the
hydrogen donor and it leads to the major product. The lowest energy TS
that leads to the minor product involves the carboxylic acid as the
donor. The computed ee’s for Reactions 5-7 are in very good, if not
excellent, agreement with the experimental values. The study should spur
further activity in which one might tune the stereoselectivity by using
catalysts with multiple binding opportunities.
References
(1) List, B.; Lerner, R. A.; Barbas, C. F., III; "Proline-Catalyzed Direct Asymmetric Aldol Reactions,"
J. Am. Chem. Soc.,
2000,
122, 2395-2396, DOI:
10.1021/ja994280y.
(2) Yang, H.; Wong, M. W. "(
S)-Proline-catalyzed nitro-Michael reactions: towards a better understanding of the catalytic mechanism and enantioselectivity,"
Org. Biomol. Chem.,
2012,
10, 3229-3235, DOI:
10.1039/C2OB06993H
(3) Lam, Y.-h.; Houk, K. N.; Scheffler, U.; Mahrwald, R.
"Stereoselectivities of Histidine-Catalyzed Asymmetric Aldol Additions
and Contrasts with Proline Catalysis: A Quantum Mechanical Analysis,"
J. Am. Chem. Soc. 2012,
134, 6286-6295, DOI:
10.1021/ja2118392

This work is licensed under a
Creative Commons Attribution-NoDerivs 3.0 Unported License.