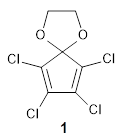
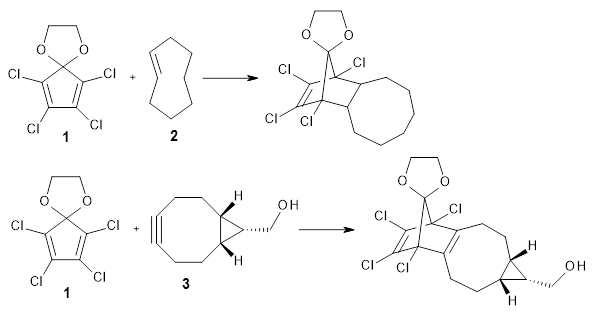
Robin J. Shannon, Emilio Martínez-Núñez, Dmitrii V. Shalashilin, David R. Glowacki (2021)
Highlighted by Jan Jensen
ChemDyME couples metadynamics statistical rate theory to automatically find kinetically important reactions and then solve the time evolution of the species in the evolving network.
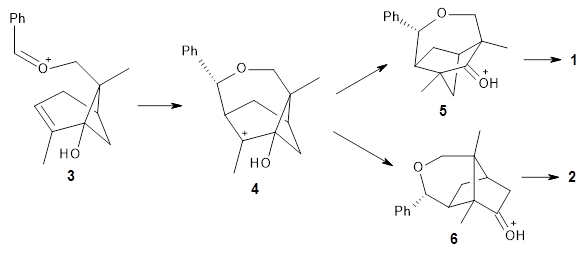
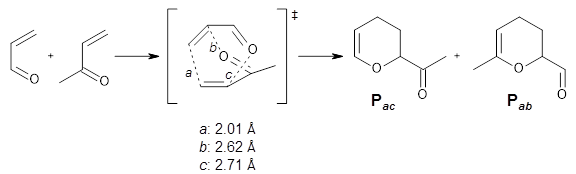
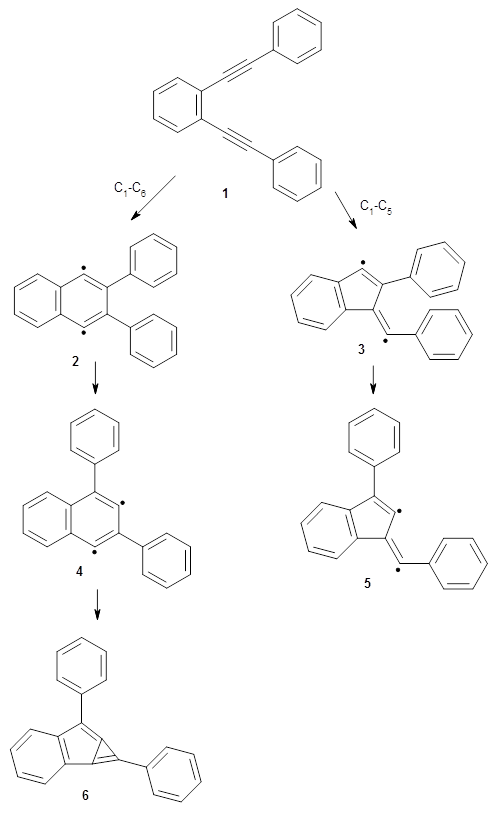
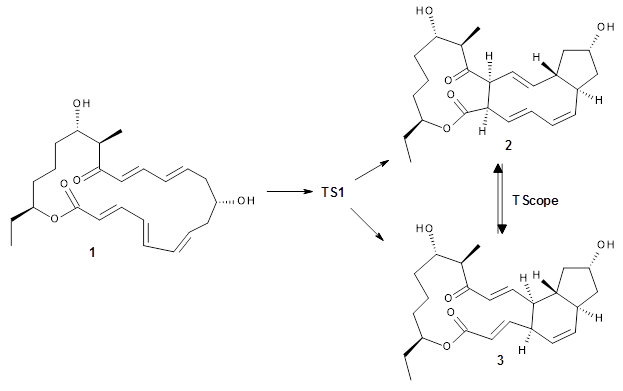
There are three steps as shown in the figure above:
1. Molecular Dynamics (MD) where semiempirical metadynamics simulations are used to identify products that are likely to be connected to the reactant by low barriers. Specifically the boxed MD (BXD) metadynamics method where an extra term to the atomic velocities to steer the MD away from previously explored regions of configuration space. The MD stops when changes in atomic connectivity is detected.
2. Optimisation and Refinement (OR) where the products structures are optimised an the TSs to the reactant are located at a higher level of theory. The initial guess for the TS geometry is the first structure in the trajectory where the atomic connectivity changes. If that approach fails a spline-based reaction path method is used. The TSs are verified by IRCs.
3. Master Equation (ME) where the set of coupled kinetic equations are solved numerically. As the reaction network grows this can become computationally demanding, which is a problem when it is done on-the-fly. The authors therefore employ the Boxed Molecular Kinetics approach to speed things up.
These steps are then repeated using the kinetically most accessible product (identified by the ME step) as the reactant. The entire procedure is then repeated until a desired maximum reaction time is reached.
The authors test the procedure on two well studies systems and show that the procedure indeed identifies the most important reactions in the reaction network.
Disclaimer: My group has developed a similar approach for the first two steps.

This work is licensed under a Creative Commons Attribution 4.0 International License.